import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
# ----------------------------------------
i_downloadData = 1 # Indicator flag whether you want to freshly download the
# data
# ----------------------------------------
if i_downloadData == 1:
urlBase = 'https://raw.githubusercontent.com/CSSEGISandData/COVID-19/master/csse_covid_19_data/csse_covid_19_time_series/'
urlConf = urlBase + 'time_series_covid19_confirmed_global.csv'
urlDead = urlBase + 'time_series_covid19_deaths_global.csv'
urlRec = urlBase + 'time_series_covid19_recovered_global.csv'
# Download and Save
dfConf = pd.read_csv(urlConf, on_bad_lines='skip')
#dfConf.to_pickle('CoronaConfirmed')
dfDead = pd.read_csv(urlDead, on_bad_lines='skip')
#dfDead.to_pickle('CoronaDeath')
dfRec = pd.read_csv(urlRec, on_bad_lines='skip')
#dfRec.to_pickle('CoronaRecovered')13 Working with Data III: Maps and Geospatial Data
In this chapter we introduce a case study using Corona virus data. We will track infection rates and plot figures using the latest data tracking the spread of the corona virus.
13.1 Importing Data
We will be working with data from the Johns Hopkins Whiting School of Engineering, Center for Systems Science and Engineering. Their Github portal is at: https://github.com/CSSEGISandData
This is the data repository for the 2019 Novel Coronavirus Visual Dashboard operated by the Johns Hopkins University Center for Systems Science and Engineering (JHU CSSE). Also, Supported by ESRI Living Atlas Team and the Johns Hopkins University Applied Physics Lab (JHU APL).
You can find their dashboard with all the visual information under this link: Dashboard
We will use their data (it's updated twice daily) and make our own graphs. We first need to import the data from their website. We can simply do this with a Pandas function .read_csv().
If you want to locally store the data and not download the data everytime you run your script file you could save the data first with:
# Save data locally on harddrive
dfConf.to_pickle(\'CoronaConfirmed\') dfDead.to_pickle(\'CoronaDeath\')
dfRec.to_pickle(\'CoronaRecovered\')And then simply read it from your harddisk using:
# Read data from harddrive
dfConf = pd.read_pickle(\'CoronaConfirmed\') dfDead =
pd.read_pickle(\'CoronaDeath\') dfRec=
pd.read_pickle(\'CoronaRecovered\')You would then of course have to "outcomment" the webreading section above or set the i_downloadData flag equal to zero so that the downloading part gets skipped.
Instead of saving the data to a file I will simply assign the imported data into a new dataframe that I am not going to manipulate.
dfConf_orig = dfConf.copy()
dfDead_orig = dfDead.copy()
dfRec_orig = dfRec.copy()For each application I will then copy the original data from the _orig dataframes.
Have a careful look at the data. Use the Variable Explorer tab in Spyder to investigate the dataframe. The nature of your data is basically an observation over time of confirmed corona virus infections by Province/State as the "smallest" geographical denominator. You also know which country the Pronvince/State belongs to (you see this in the second column) and then you also have the Latitude/Longitude coordinates of the Province/State from which the corona cases are reported from. This is followed by daily observations from this Province/State.
13.2 Plotting Cases of Infections
I first copy the original data into new dataframes because I want to keep the raw data intact and untouched in case I want to come back to it later, which we will!
dfConf = dfConf_orig.copy()
dfDead = dfDead_orig.copy()
dfRec = dfRec_orig.copy()We next add a column with the sum of all the confirmed coronavirus cases for each Province/State. In other words, we sum up all the columns of the time series of cases which starts in column five, so that we go from [4:] to the end.
dfConf['Confirmed']=dfConf.iloc[:,-1]
dfDead['Dead']=dfDead.iloc[:,-1]
dfRec['Recovered']=dfRec.iloc[:,-1]We then drop the entire time series and only keep the overall sum of cases. We are not interested in the single day observations for this first summary graph.
dfConf.drop(dfConf.iloc[:, 4:-2], inplace = True, axis = 1)
dfDead.drop(dfDead.iloc[:, 4:-2], inplace = True, axis = 1)
dfRec.drop(dfRec.iloc[:, 4:-2], inplace = True, axis = 1)We next merge the three dataframes together by Province/State, Country/Region, Lat, and Long variables so that we have the sum of all confirmed infection cases, the sum of all corona virus associated deaths, and the sum of all the recovered cases for each Province/State in the same dataframe.
dftemp = pd.merge(dfConf, dfDead, \
on=['Province/State', 'Country/Region','Lat','Long'], how='inner')
df = pd.merge(dftemp, dfRec, \
on=['Province/State', 'Country/Region','Lat','Long'], how='inner')We have now one dataframe with the sum of all confirmed coronavirus cases, the sum of all deaths due to corona virus, as well as the sum of all recorded recoveries from a coronavirus infection.
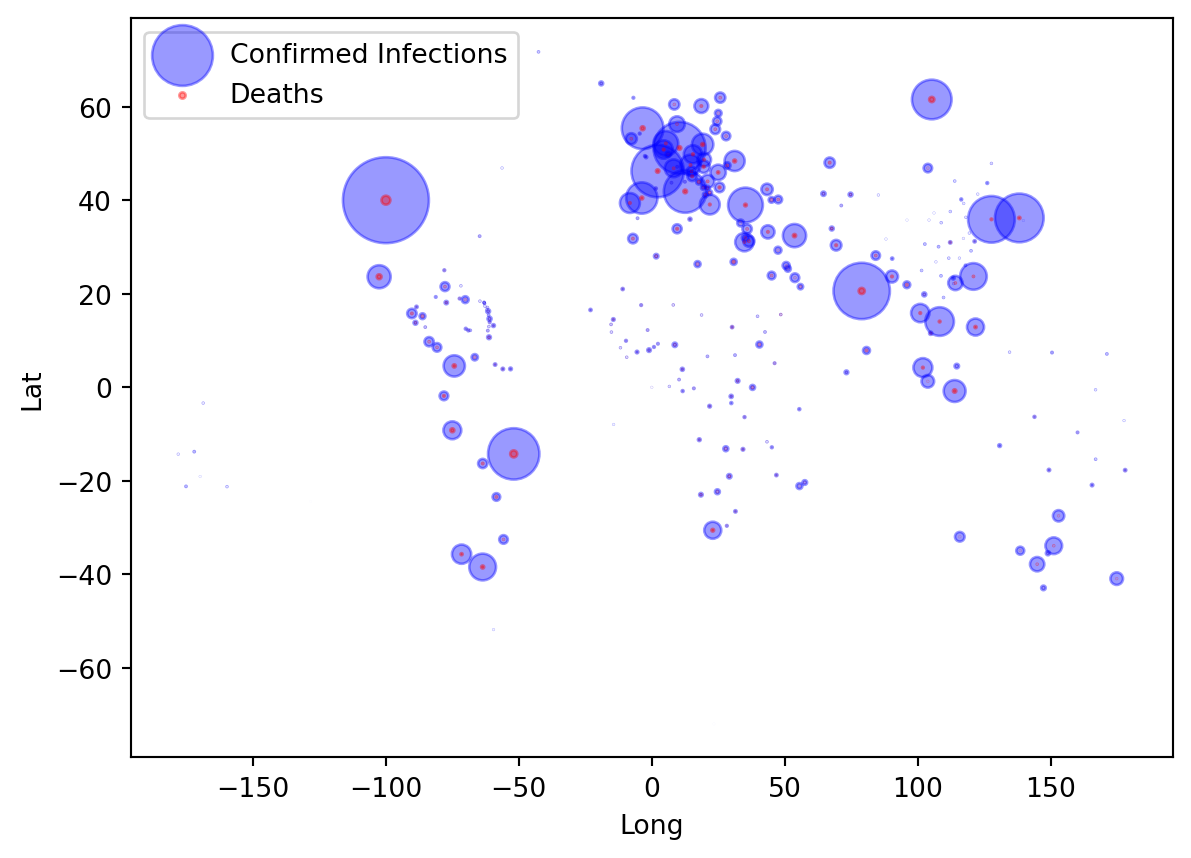
We next plot the infection cases by their latitude and longitude of the province/state where they were recorded. We plot circles and use the number of cases per 1000 as circle size. The larger the circle in the plot, the more cases have been recorded for the Latitude/Longitude coordinate.
ax = df.plot(kind="scatter", x="Long", y="Lat", alpha=0.4,
s=df["Confirmed"]/100000, label="Confirmed Infections", color = "Blue")
df.plot(kind="scatter", x="Long", y="Lat", alpha=0.4,
s=df["Dead"]/100000, label="Deaths", color="Red", ax=ax)
plt.legend()
plt.show()From this graph you can already see the outline of countries. However, it would be better if we could superimpose the information onto a real map of the world. We do this next.
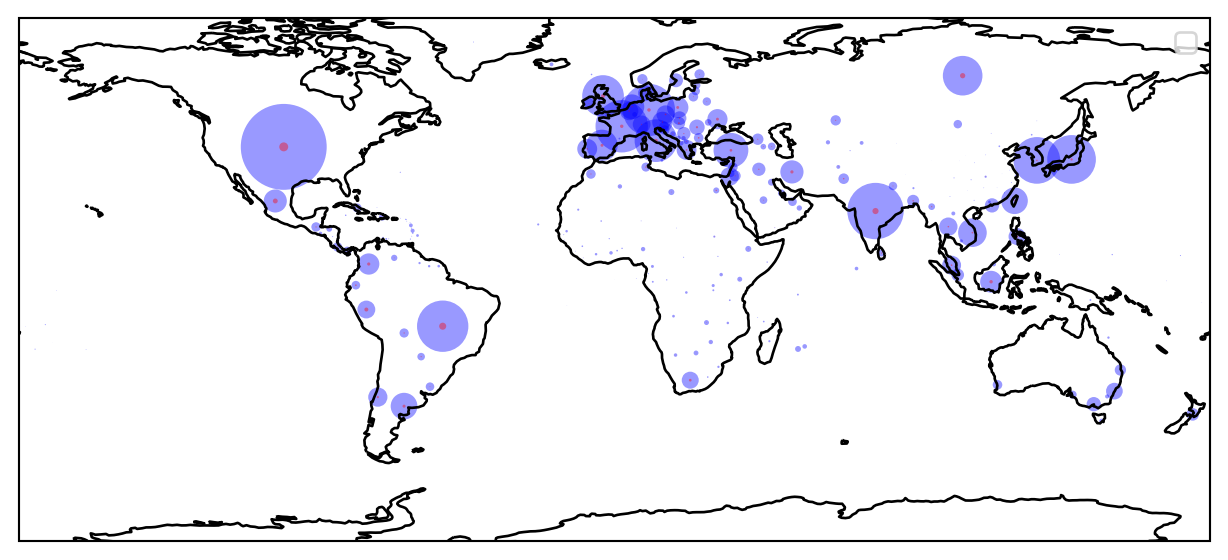
13.3 Plotting Cases of Infections on a Map
We next use the Cartopy library to plot the same information superimposed on a worl map.
You will need to install the cartopy library via the command line. Open a command line terminal and type:
conda install cartopyFollowed by enter, then hit y for yes when it prompts you. This should install the cartopy library.
We will next import the cartopy library and superimpose the same plot on a World map in Figure 13.2 .
import cartopy.crs as ccrsfig = plt.figure(figsize=(8, 10))
ax = plt.axes(projection=ccrs.PlateCarree())
ax.coastlines()
plt.scatter(df['Long'].values, df['Lat'].values, transform=ccrs.PlateCarree(), \
label=None, s=df["Confirmed"]/100000, c="Blue", linewidth=0, alpha=0.4)
plt.scatter(df['Long'].values, df['Lat'].values, transform=ccrs.PlateCarree(), \
label=None, s=df["Dead"]/100000, c="Red", linewidth=0, alpha=0.4)
plt.legend()
plt.show()No artists with labels found to put in legend. Note that artists whose label start with an underscore are ignored when legend() is called with no argument.You now have a nice plot of the world map and the corona cases superimposed on it.
13.4 Plotting Time Series of Cases
We now go back to the original dataframe with the time series data of confirmed coronavirus cases. We then drop some variables that we do not need, such as Province/State, Lat, and Long.
# Here is the original data again
dfConf = dfConf_orig.copy()
dfDead = dfDead_orig.copy()
dfRec = dfRec_orig.copy()Now pick the confirmed cases and drop some columns
dfConf_t = dfConf
dfConf_t = dfConf_t.drop(columns = ['Province/State', 'Lat', 'Long'])Let us have a look at the first three rows to see how our raw data look like.
print(dfConf_t[0:3]) Country/Region 1/22/20 1/23/20 1/24/20 1/25/20 1/26/20 1/27/20 \
0 Afghanistan 0 0 0 0 0 0
1 Albania 0 0 0 0 0 0
2 Algeria 0 0 0 0 0 0
1/28/20 1/29/20 1/30/20 ... 2/28/23 3/1/23 3/2/23 3/3/23 3/4/23 \
0 0 0 0 ... 209322 209340 209358 209362 209369
1 0 0 0 ... 334391 334408 334408 334427 334427
2 0 0 0 ... 271441 271448 271463 271469 271469
3/5/23 3/6/23 3/7/23 3/8/23 3/9/23
0 209390 209406 209436 209451 209451
1 334427 334427 334427 334443 334457
2 271477 271477 271490 271494 271496
[3 rows x 1144 columns]We next add up all the cases for each day by Country/Region using the groupby() function that comes with Pandas.
dfConf_t = dfConf_t.groupby('Country/Region').sum()And again, let us peek at the data real quick.
print(dfConf_t[0:3]) 1/22/20 1/23/20 1/24/20 1/25/20 1/26/20 1/27/20 1/28/20 \
Country/Region
Afghanistan 0 0 0 0 0 0 0
Albania 0 0 0 0 0 0 0
Algeria 0 0 0 0 0 0 0
1/29/20 1/30/20 1/31/20 ... 2/28/23 3/1/23 3/2/23 \
Country/Region ...
Afghanistan 0 0 0 ... 209322 209340 209358
Albania 0 0 0 ... 334391 334408 334408
Algeria 0 0 0 ... 271441 271448 271463
3/3/23 3/4/23 3/5/23 3/6/23 3/7/23 3/8/23 3/9/23
Country/Region
Afghanistan 209362 209369 209390 209406 209436 209451 209451
Albania 334427 334427 334427 334427 334427 334443 334457
Algeria 271469 271469 271477 271477 271490 271494 271496
[3 rows x 1143 columns]We now transpose the dataframe because we want the time observations as rows and not columns.
dfConf_t = dfConf_t.T
print(dfConf_t[0:3])Country/Region Afghanistan Albania Algeria Andorra Angola Antarctica \
1/22/20 0 0 0 0 0 0
1/23/20 0 0 0 0 0 0
1/24/20 0 0 0 0 0 0
Country/Region Antigua and Barbuda Argentina Armenia Australia ... \
1/22/20 0 0 0 0 ...
1/23/20 0 0 0 0 ...
1/24/20 0 0 0 0 ...
Country/Region Uruguay Uzbekistan Vanuatu Venezuela Vietnam \
1/22/20 0 0 0 0 0
1/23/20 0 0 0 0 2
1/24/20 0 0 0 0 2
Country/Region West Bank and Gaza Winter Olympics 2022 Yemen Zambia \
1/22/20 0 0 0 0
1/23/20 0 0 0 0
1/24/20 0 0 0 0
Country/Region Zimbabwe
1/22/20 0
1/23/20 0
1/24/20 0
[3 rows x 201 columns]We are now ready to convert the index of the dataframe into a date index so that we can use the built in time series commands in Pandas.
# Converting the index as date
dfConf_t.index = pd.to_datetime(dfConf_t.index)print(dfConf_t[0:3])Country/Region Afghanistan Albania Algeria Andorra Angola Antarctica \
2020-01-22 0 0 0 0 0 0
2020-01-23 0 0 0 0 0 0
2020-01-24 0 0 0 0 0 0
Country/Region Antigua and Barbuda Argentina Armenia Australia ... \
2020-01-22 0 0 0 0 ...
2020-01-23 0 0 0 0 ...
2020-01-24 0 0 0 0 ...
Country/Region Uruguay Uzbekistan Vanuatu Venezuela Vietnam \
2020-01-22 0 0 0 0 0
2020-01-23 0 0 0 0 2
2020-01-24 0 0 0 0 2
Country/Region West Bank and Gaza Winter Olympics 2022 Yemen Zambia \
2020-01-22 0 0 0 0
2020-01-23 0 0 0 0
2020-01-24 0 0 0 0
Country/Region Zimbabwe
2020-01-22 0
2020-01-23 0
2020-01-24 0
[3 rows x 201 columns]Drop the last observation because it is an empty row.
if np.sum(dfConf_t.iloc[-1,:].values) > 0:
# Do nothing
print('Data complete.')
else:
# If data is not there yet, drop last row
dfConf_t = dfConf_t[:-1]Data complete.We are now ready to plot the time series for different countries. We can choose the number of days we want to plot. Here we choose the most recent 700 observations.
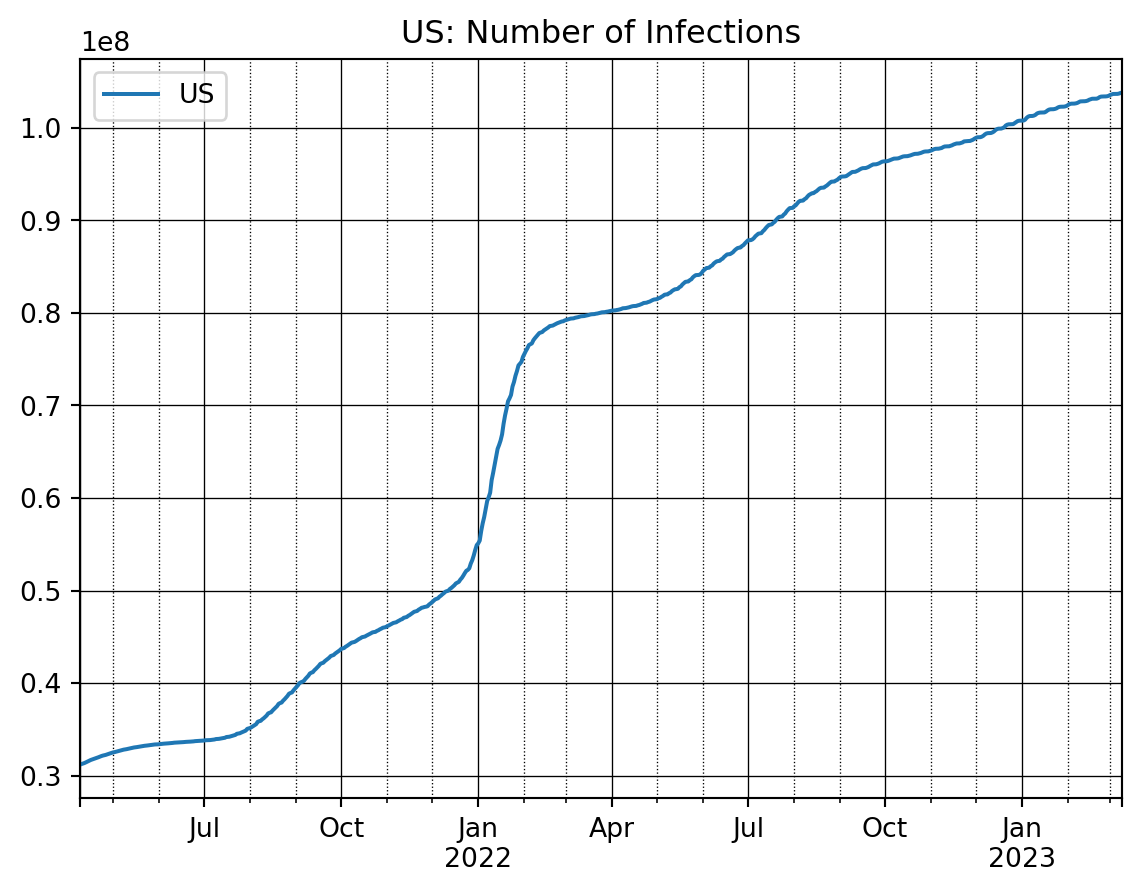
nrObs = -700 # Just plot the recent 700 obs (days)
ax = dfConf_t['US'].iloc[nrObs:].plot()
ax.set_title('US: Number of Infections')
# Customize the major grid
ax.grid(which='major', linestyle='-', linewidth='0.5', color='Black')
# Customize the minor grid
ax.grid(which='minor', linestyle=':', linewidth='0.5', color='black')
plt.legend()
plt.show()In order to plot multiple countries into a single graph, I first make a list of countries and then run a loop over this list and invoke the plot command. Otherwise we would have a lot of repeat code which is bad programming style.
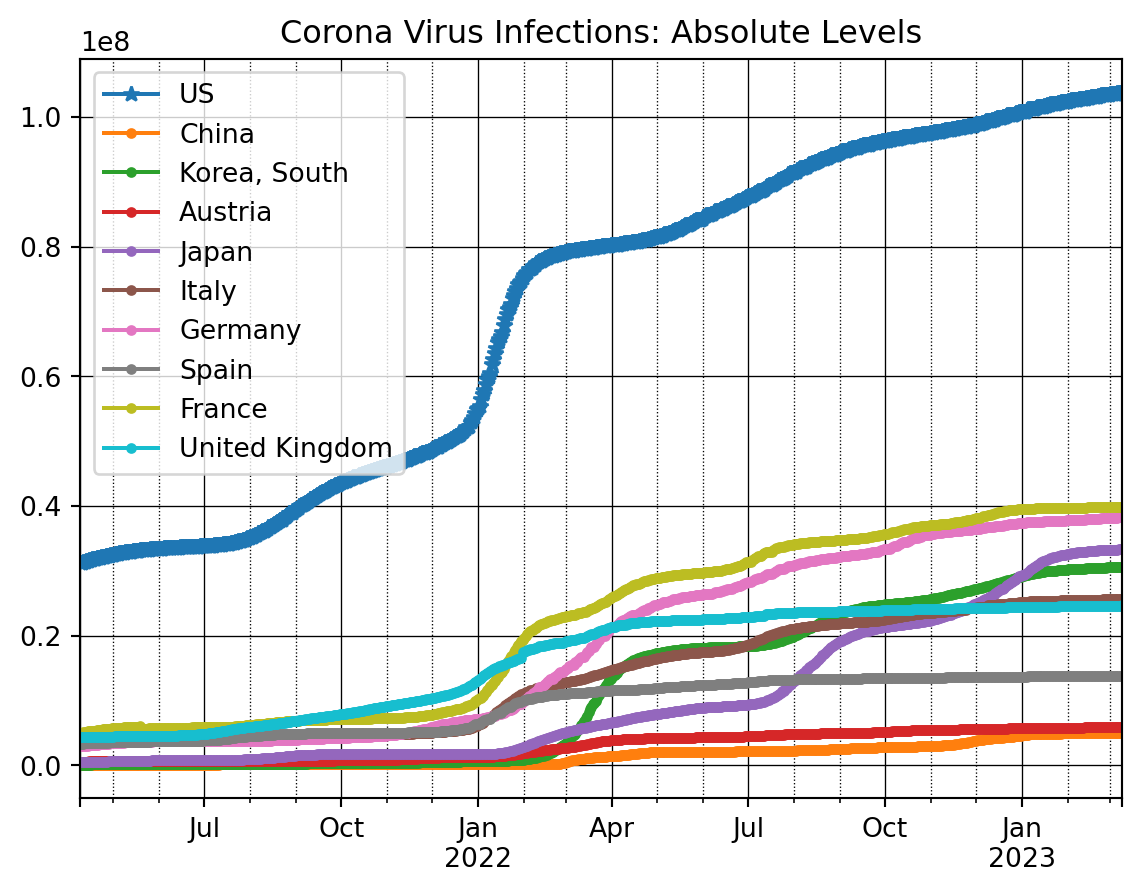
# Plot
countryList = ['US', 'China', 'Korea, South', 'Austria', 'Japan', 'Italy', \
'Germany', 'Spain', 'France', 'United Kingdom']
# Shorter list for alternative graph
# countryList = ['US', 'Korea, South', 'Austria', 'Japan', \
# 'Germany', 'Spain', 'France', 'United Kingdom']
nrObs = -700
ax = dfConf_t[countryList[0]].iloc[nrObs:].plot(marker = '*')
for x in countryList[1:]:
dfConf_t[x].iloc[nrObs:].plot(ax=ax, marker = '.')
ax.set_title('Corona Virus Infections: Absolute Levels')
# Customize the major grid
ax.grid(which='major', linestyle='-', linewidth='0.5', color='Black')
# Customize the minor grid
ax.grid(which='minor', linestyle=':', linewidth='0.5', color='black')
plt.legend()
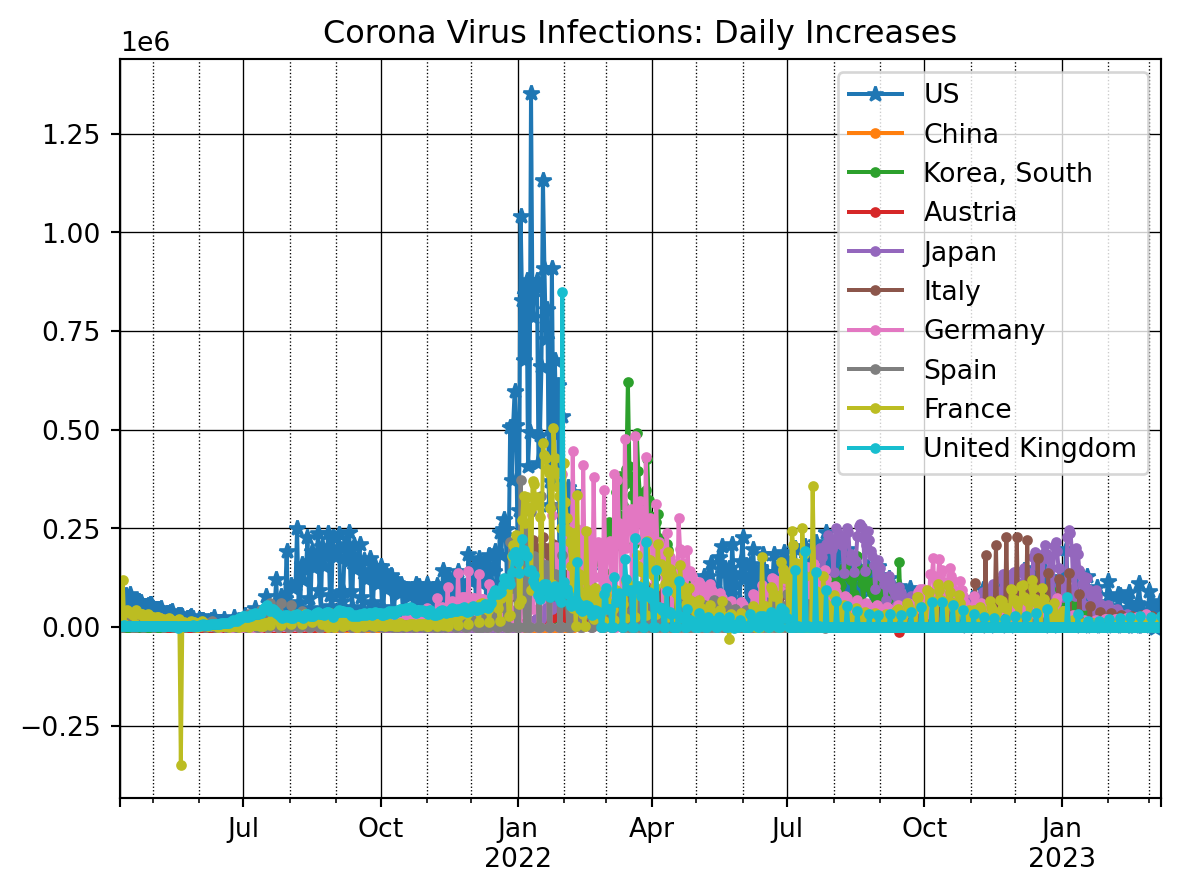
plt.show()We next investigate the changes in the numbers from one day to the next using the diff() function. It basically subtracts consecutive observations from each other, i.e., it takes the number of infections from day t and subtracts the number of infections from the prior day t-1.
ax = dfConf_t[countryList[0]].iloc[nrObs:].diff().plot(marker = '*')
for x in countryList[1:]:
dfConf_t[x].iloc[nrObs:].diff().plot(ax=ax, marker = '.')
ax.set_title('Corona Virus Infections: Daily Increases')
# Customize the major grid
ax.grid(which='major', linestyle='-', linewidth='0.5', color='Black')
# Customize the minor grid
ax.grid(which='minor', linestyle=':', linewidth='0.5', color='black')
plt.legend()
plt.show()13.5 Plotting Time Series of Corona Virus Deaths
We again need to transform the data into a time series dataframe first.
# Death Rates
dfDead_t = dfDead
dfDead_t = dfDead_t.drop(columns = ['Province/State', 'Lat', 'Long'])
dfDead_t = dfDead_t.groupby('Country/Region').sum()
dfDead_t = dfDead_t.T
# Converting the index as date
dfDead_t.index = pd.to_datetime(dfDead_t.index)
if np.sum(dfDead_t.iloc[-1,:].values) > 0:
# Do nothing
print('Data complete.')
else:
# If data is not there yet, drop last row
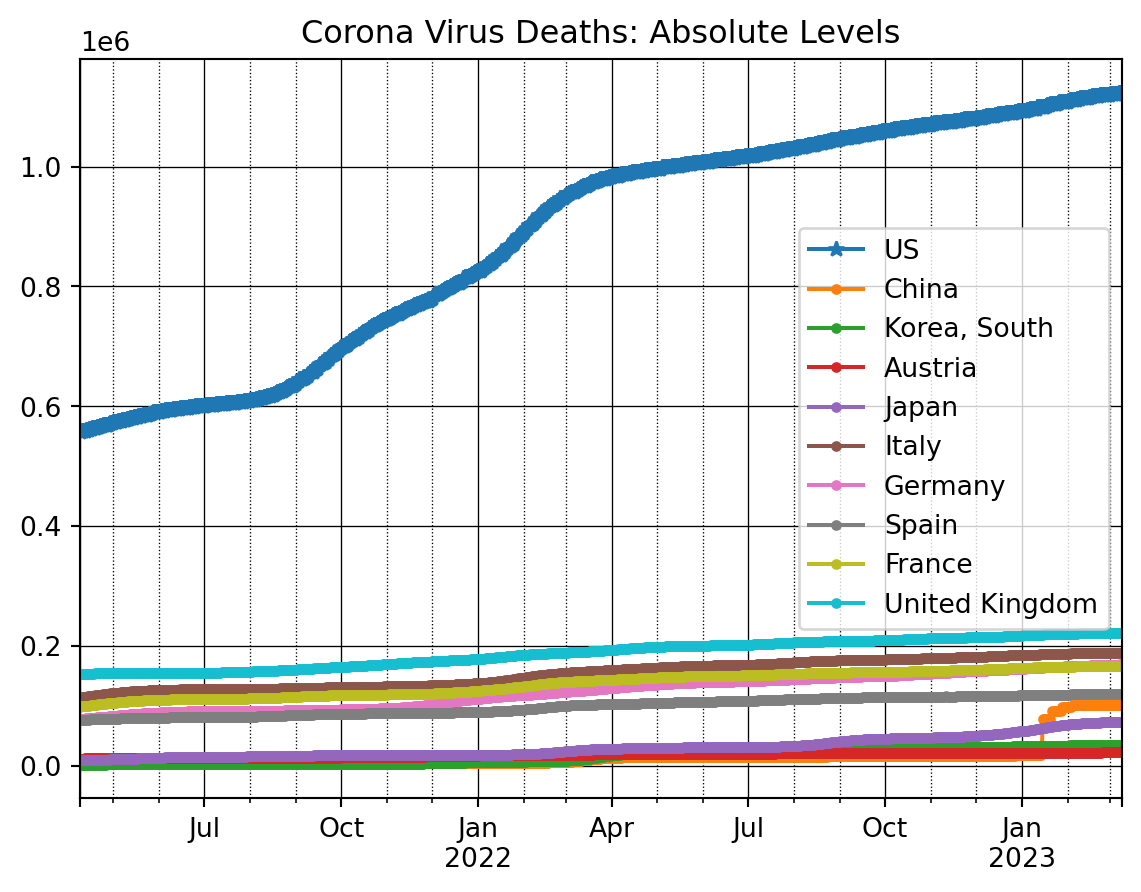
dfDead_t = dfDead_t[:-1]Data complete.And we can now plot the information. We again start with the absolute levels.
ax = dfDead_t[countryList[0]].iloc[nrObs:].plot(marker = '*')
for x in countryList[1:]:
dfDead_t[x].iloc[nrObs:].plot(ax=ax, marker = '.')
ax.set_title('Corona Virus Deaths: Absolute Levels')
# Customize the major grid
ax.grid(which='major', linestyle='-', linewidth='0.5', color='Black')
# Customize the minor grid
ax.grid(which='minor', linestyle=':', linewidth='0.5', color='black')
plt.legend()
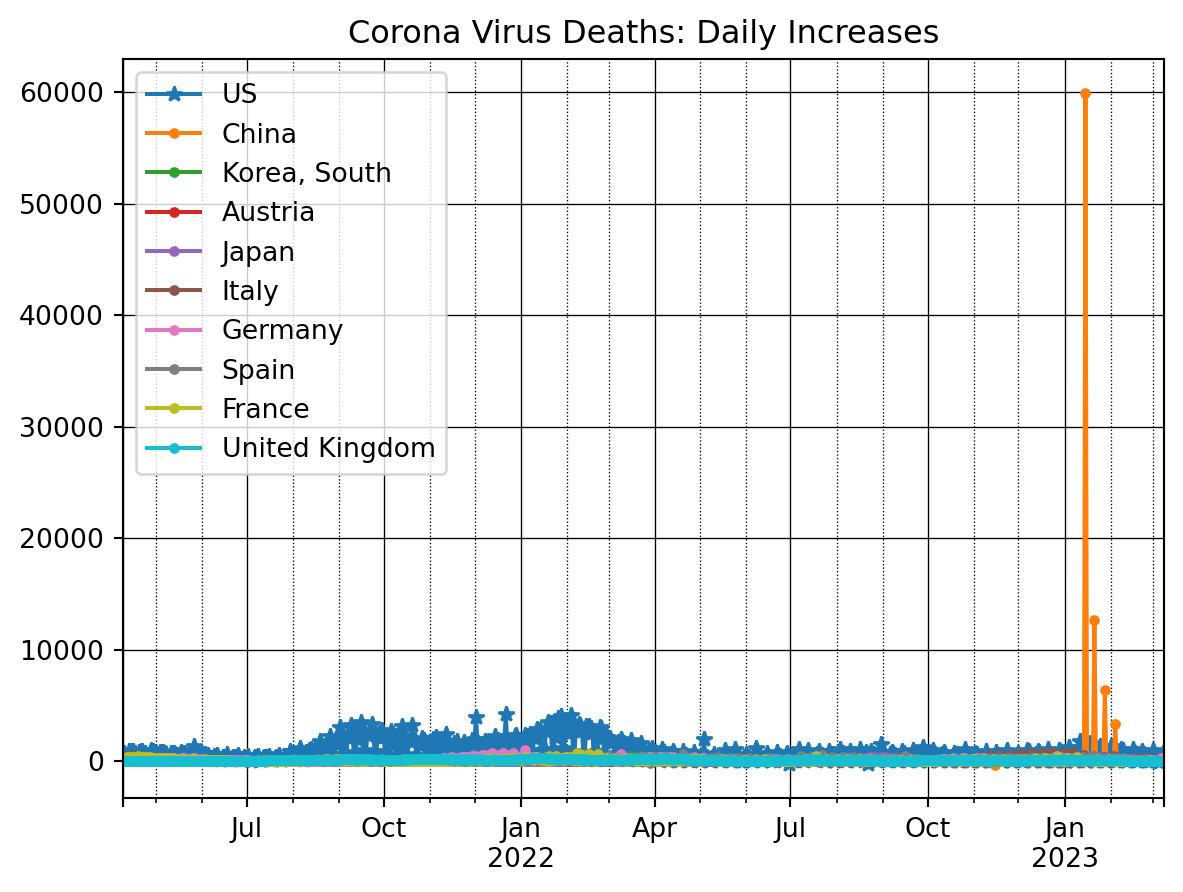
plt.show()The daily changes in number of deaths can be plotted as follows
ax = dfDead_t[countryList[0]].iloc[nrObs:].diff().plot(marker = '*')
for x in countryList[1:]:
dfDead_t[x].iloc[nrObs:].diff().plot(ax=ax, marker = '.')
ax.set_title('Corona Virus Deaths: Daily Increases')
# Customize the major grid
ax.grid(which='major', linestyle='-', linewidth='0.5', color='Black')
# Customize the minor grid
ax.grid(which='minor', linestyle=':', linewidth='0.5', color='black')
plt.legend()
plt.show()13.6 Key Concepts and Summary
13.7 Self-Check Questions
- Generate graphs that track the corona virus infection rates for all 50 US states
- Generate graphs that show the daily change of number of infections for all 50 US states.